In the previous post, we have seen the basics of nucleic
acids and the native form of DNA i.e.; B-DNA in details.
There are various other forms which DNA can assume. In this post, I will discuss some of the other forms of DNA (i.e.; A-DNA and Z-DNA) and another type of nucleic acid - RNA.
Starting the post with the A-form of DNA.
The following are certain characteristics of A-DNA:
i. This form of DNA exists when the relative humidity
is around 75%.
ii. The A-DNA is also
right-handed helix like B-DNA.
iii. A-DNA is shorter and wider than B-DNA.
iii. A-DNA is shorter and wider than B-DNA.
iv. The diameter
of A-DNA is 23Å as against 20Å in B-DNA.
v. There are 11 base pairs per turn (more
than B-DNA which has 10 base pairs per turn).
vi. The rise per turn is 28Å.
vii. Thus,
if we calculate, rise per base pair (28Å/11base pairs), we get 2.6Å.
viii. The base pairs are not
perpendicular to the helical axis instead they are tilted 20˚ w.r.t. helical
axis.
ix. Just like B-DNA, A-DNA also has major groove and minor groove; however, the former is very deep and latter is very shallow as can be seen in the adjacent diagram.
Does A-DNA have any
biological importance? Yes. A-DNA is observed in some biological context as follows.
A ̴ 3 base pair segment of A-DNA is present at the active site of
DNA polymerase.
Also, gram positive bacteria undergoing sporulation contain a high proportion (20%) of small acid soluble-spore proteins (SASPs). Some of these SASPs induce B-DNA to assume the A-form of DNA (atleast in vitro).
Another instance where A-form of DNA is present is during RNA-DNA hybrids. RNA-DNA hybrids do occur in the cell at different junctions for example, during replication (where replication is initiated by RNA primer) and also during transcription (where RNA is made on DNA template). These RNA-DNA hybrids cannot take up the B-form of DNA (because of the 2' oxygen of RNA) and hence, they resemble A-form of DNA.
Also, gram positive bacteria undergoing sporulation contain a high proportion (20%) of small acid soluble-spore proteins (SASPs). Some of these SASPs induce B-DNA to assume the A-form of DNA (atleast in vitro).
Another instance where A-form of DNA is present is during RNA-DNA hybrids. RNA-DNA hybrids do occur in the cell at different junctions for example, during replication (where replication is initiated by RNA primer) and also during transcription (where RNA is made on DNA template). These RNA-DNA hybrids cannot take up the B-form of DNA (because of the 2' oxygen of RNA) and hence, they resemble A-form of DNA.
Another form of DNA is Z-DNA and is called so because of its zigzag pattern of the phosphate
backbone. The following are some of the characteristics:
i. The main characteristic is that Z-DNA is a left-handed double helix.
ii. The formation of Z-DNA is base sequence dependent (composition
dependent). Only alternating purine and pyrimidine polymers can form Z-DNA.
Thus, this zigzag forms the repeating unit of Z-DNA which is a dinucleotide.
iii. The
right handed DNA can be transited to left handed DNA (Z-DNA) in solutions that include high
ionic strength, hydrophobic solvents, presence of certain trivalent cations or
covalent modification with bulky alkylating agents.
iv. It contains approximately 12 base pairs per turn of the helix.
v. Rise per turn of the helix is 44Å.
vi. The helix rises by 2.7Å per base pair.
vii. The
base pairs are perpendicular of the helix (just like that of B-DNA).
viii. Regarding the grooves in Z-DNA, it possesses only a minor groove.
There is no discernible major groove and the minor groove is extremely deep and
narrow.
The following table will give you an idea of major structural differences between three major forms of DNA (B-DNA, A-DNA and Z-DNA).
Ribonucleic Acid (RNA):
Coming to another type of nucleic acid which is RNA. As we have seen the major difference between
RNA and DNA in previous post, here I will discuss about its types and structure.
Types of RNA:
There are various kinds of RNAs. Here, the RNAs have been classified into two major groups as coding RNAs and non-coding RNAs.
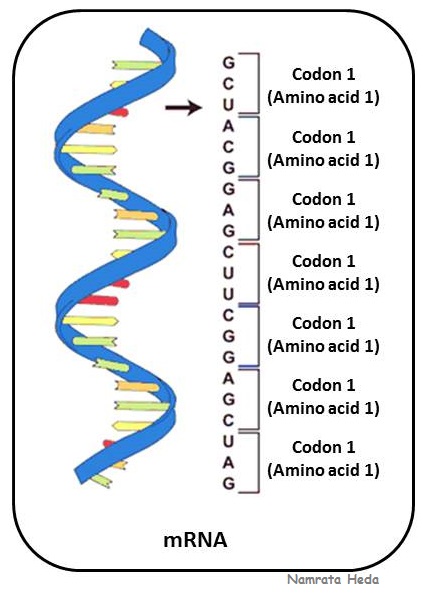
mRNA (messenger RNA) is
coding RNA that is involved in the process of translation in the cell. It is
called messenger as it carries information from DNA to the ribosome which is
the site of protein synthesis. The coding sequence of mRNA determines the amino
acid sequence in the protein that is produced.
 b. Non-coding RNAs:
b. Non-coding RNAs:Most of the RNAs do not code for any proteins and these types of RNA fall under another group as non-coding RNAs (ncRNAs). These so called non-coding RNAs can be encoded by their own genes. The most prominent examples are rRNA (ribosomal RNA) and tRNA (transfer RNA), both of which are involved in the process of translation.
Some other examples of noncoding RNAs are those involved in
gene regulation and RNA processing. There are also some non coding RNAs that
are able to catalyse chemical reactions such as cutting and ligating other RNA molecules and catalysis of peptide bond formation in
ribosome. Such catalytic RNAs are called ribozymes.
The structure of RNA is described as follows:
Primary Structure of RNA:
Primary structure of RNA is somewhat similar to that of DNA; the only difference being the sugar (ribose) component which has an additional hydroxyl group at 2' carbon and thymine is replaced with uracil (see figure on the left).
Note: The hydroxyl group on the 2' carbon atom of ribose make it more labile as compared to DNA and it provides a chemically reactive group. Because of this lability, RNA is cleaved into mononucleotides by alkaline solution and DNA is not.
Primary structure of RNA is somewhat similar to that of DNA; the only difference being the sugar (ribose) component which has an additional hydroxyl group at 2' carbon and thymine is replaced with uracil (see figure on the left).
Note: The hydroxyl group on the 2' carbon atom of ribose make it more labile as compared to DNA and it provides a chemically reactive group. Because of this lability, RNA is cleaved into mononucleotides by alkaline solution and DNA is not.
RNA is a polynucleotide chain that can be double stranded or
single stranded, linear or cicular.
Secondary Structure of RNA:
 There are various types of RNA
exhibiting different conformations. The simplest secondary structures in single
stranded RNAs are formed by pairing of complementary bases. Two major forms of secondary structures are hairpins and stem loops.
There are various types of RNA
exhibiting different conformations. The simplest secondary structures in single
stranded RNAs are formed by pairing of complementary bases. Two major forms of secondary structures are hairpins and stem loops.‘Hairpins’ are formed by pairing within approximately 5-10 nucleotides of each other whereas in stem loop, there is pairing of bases that are separated by approximately 50 to several hundreds of nucleotides.
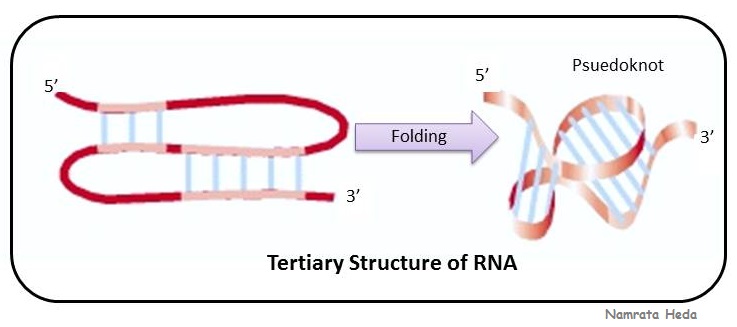
These simple folds (hairpin and
stem-loops) can cooperate to form more complicated tertiary structures, and one of such structure is 'pseudoknot' as can be seen in the adjacent figure.
tRNA is a well-defined example of tertiary structure of RNA. tRNA is a type of RNA that is involved in translation process of protein synthesis. They have a L-shaped 3D structure that allows them to fit into the P and A-sites of the ribosome. The figure depicted as tRNA above under non-coding RNAs is the three-dimensional structure of tRNA.
Here, we complete the nucleic acids topic. Any comments or doubts are welcome.!
tRNA is a well-defined example of tertiary structure of RNA. tRNA is a type of RNA that is involved in translation process of protein synthesis. They have a L-shaped 3D structure that allows them to fit into the P and A-sites of the ribosome. The figure depicted as tRNA above under non-coding RNAs is the three-dimensional structure of tRNA.
Here, we complete the nucleic acids topic. Any comments or doubts are welcome.!